Joseph W. Francis
Associate Professor of Biology
Cedarville College, Ohio
francisj@cedarville.edu
The smallest living building block of life, the cell, is enormously complex, and a great number of its mechanisms are irreducibly complex. Few theories have been proposed explaining how irreducibly complex mechanisms could have evolved by Darwinian natural selection. It could be argued that given enough time a simple reproducing population of living “protocells” could have provided a format for the evolution of complex mechanisms. However, even in “simple” bacteria, the most basic cell functions display irreducibly complex mechanisms—for instance, cell division. This article considers the origin of an irreducibly complex cell division apparatus and contrasts protocell theory with intelligent design theory.
“The only life we know for certain is cellular.....”
— Harold J. Morowitz
Protocell theory is a popular theory often proposed to explain how biochemical complexity arose in living cells by completely natural, evolutionary processes. The theory postulates that the complex cells we observe today evolved gradually from simpler protocells via natural selection. For example, Harold Morowitz has suggested that the original protocells were unstable and prone to self-destruction; but through the continual formation of billions of protocells over millions of years, eventually a stable, more advanced, protocell formed (Morowitz, 1992).
In contrast to protocell theory, intelligent design theory postulates that some biochemical mechanisms within cells are irreducibly complex, which implies that they are not products of any gradual, naturalistic process of formation. For example, many cell mechanisms resemble preassembled machines containing interdependent parts that work together to perform a cell function. Since all the co-dependent parts must be present before the mechanism is capable of function at all, it is unlikely that the mechanism evolved by any gradual process, but instead appears to have been designed.
Examples of irreducibly complex biological systems have been documented by biochemist Michael Behe in Darwin’s Black Box (Behe, 1996). Behe defines irreducible complexity as “a single system of several well matched, interacting parts that contribute to the basic function, where the removal of any one of the parts causes the system to effectively cease functioning.” As one of several examples, Behe cites the bacterial flagellum, a whip-like cell appendage that has a complex motor apparatus at its base. How did such a complex structure evolve in what is often described as a “simple” cell? Protocell theorists propose that the bacterial flagellum evolved gradually by natural selection (Loomis, 1984), implying that such complex mechanisms are late developments in the evolution of bacteria and would not have been present in early protocells.
If, however, the protocell is a self-reproducing cell, as the theory suggests, then several essential cell functions that appear to be complex in contemporary cells would have to be present even in the early protocell. For instance, a cell division mechanism would be essential even in early stages of protocell evolution. The impression given in many biology textbooks is that cell division is a simple process. However, upon close examination it becomes clear that cell division even in bacteria is a complex cellular process. This raises several questions concerning protocell theory. For instance, how did a simpler cell division mechanism function? How did it lead to the complex mechanism we observe today? Are any remnants of a simpler cell division process evident in cells today? Is the cell division process irreducibly complex?
The existence of an irreducibly complex cell division process would present two problems for protocell theory. First, it would require the theory to explain the origin of a protocell that must possess, at a minimum, an irreducibly complex cell division process for survival. Second, if a protocell is capable of surviving with a simple cell division process, how does natural selection lead to a more complex, indeed an irreducibly complex, cell division mechanism?
In considering these questions, I will examine cell division in bacteria. There is general agreement among biologists that bacteria represent a late protocell, or at least an evolutionary link between a simpler protocell and eukaryotes, based on the fact that bacteria appear to be simple cells, both morphologically and genetically, compared with eukaryotes. Bacteria are also well adapted to independent unicellular life and hostile environments, conditions postulated to exist on the early earth. However, the assumption that bacteria are simple is itself open to question, as we shall see. I will focus on one primary question: Is there a minimum set of reactions a bacterium must possess to divide and reproduce, and, if so, are these mechanisms irreducibly complex?
Let’s begin by listing the essential processes a bacterium must possess to reproduce effectively. For the sake of simplicity, we will ignore for the time being the requirement for energy in the form of ATP and its biochemical production in the cell.
The common link among all the processes mentioned above is the need for proteins. In fact, if protein synthesis is inhibited, cell division ceases (Lewin, 1997). Therefore, unless there is a source of pre-manufactured proteins that can be transported into the cell, the cell itself must contain a protein synthesis process. Even in bacteria, protein synthesis is a highly complex and regulated process, involving many proteins and machine-like protein complexes. More than 200 hundred proteins involved in this process have been identified in the bacterium Escherichia coli (E.coli) (Javor, 1998). Without this protein synthesis machinery, bacteria would not be able to divide (nor even keep the cell’s “house” in order).1
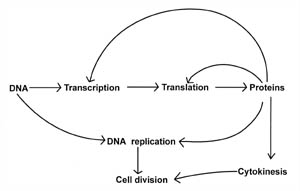
Let’s now consider our central question: Are the complex biochemical processes that control cell division representative of a minimal set of reactions that the cell requires for life, and are they irreducibly complex? There is good evidence to suggest that the process of cell division is indeed irreducibly complex, for the steps involved are interdependent and highly coordinated. For example, crucial steps such as DNA transcription require proteins (see Figure 1)—while protein synthesis in turn is dependent upon transcription. Moreover, evidence suggests that the processes involved in cell division are highly regulated and coordinated in a sequential fashion. For instance, in bacteria, cytokinesis does not proceed until DNA replication is complete, so that the DNA is precisely partitioned into the developing daughter cells. Each process itself is complex and if any one of the processes is inhibited, cell division ceases. This interdependence fits the criteria of an irreducibly complex system.

Figure 1. Relationship between some of the basic biochemical processes that are required for life and cell division. Transcription and translation are processes that are involved in making proteins by deciphering the genetic code (DNA). However, proteins are required for both the transcription and translation processes, as well as for DNA replication and cytokinesis. Note that proteins play a vital role in each cell process and serve as the major interconnecting link between each process. Cell division is a protein-dependent mechanism.
Does this arrangement also represent a minimal system that must be present in all cells, including any hypothetical protocell? Or could it have evolved gradually? Let’s consider the possible gradual derivation of the cell division apparatus in the protocell.
One of the more popular theories of protocell evolution, presented in biology textbooks, involves the encapsulation of the basic processes of biopolymer synthesis in a membrane (Cooper, 1997). It is then postulated that the protocell began to divide by a simple mechanism. In other words, it is assumed that all the cell functions required for life, perhaps even those required for cell division, were pre-manufactured and pre-functioning processes sequestered together by a cell membrane. (One barrier to cell division that the early protocell would encounter is that in an aqueous environment there is a natural physical resistance to the membrane disruption needed for cell division. For the sake of discussion, we will assume that the dividing protocell was in a membrane-disrupting environment that promoted some type of membrane blebing or stressing so that new cells could bud or pinch off the protocell.)
There are several fundamental problems with the encapsulation theory. First, how does a cytokinesis process develop before the membrane forms the cell? Cytokinesis requires a membrane-enclosed cytoplasmic space and could only develop after encapsulation. Yet in that case—if cytokinesis evolved only after encapsulation—then it would have to evolve rapidly, otherwise the cell would not reproduce and its long-term survival would be questionable. One possible postulate is that the early cytokinesis process was a much simpler process compared with the complex cytokinesis mechanism observed in bacteria today. That would imply, however, that there was very little regulation or no coordination between DNA replication and cytokinesis and other cell systems, which in turn implies that the division of the membrane and successful transfer of genetic material was haphazard and inefficient. The protocell would partition its DNA into new daughter bacteria, and then divide, by random uncoordinated processes.
Let’s examine some hypothetical protocell models that involve cell division without coordination between the processes of DNA replication, protein synthesis, and cytokinesis. To keep the model simple we will portray the early protocell as monoploid—i.e., containing one strand of DNA (though some researchers suggest that the early protocell may have been polyploid, harboring short pieces of DNA).
In contemporary bacteria, DNA replication precedes cytokinesis so that a single cell momentarily has what appear to be two copies of circular DNA; thereafter the DNAs are partitioned into two new daughter bacteria, resulting in two separate daughter bacteria with one circular DNA molecule each. If, however, in the early protocell there was no coordination between cytokinesis and DNA replication, we can predict two scenarios for how early protocell division could have occurred: either DNA replication occurred at a faster rate than cytokinesis, or, conversely, cytokinesis occurred at a faster rate than DNA replication. If cytokinesis occurred at a faster rate, the result would be the production of many anucleate, nonviable bacteria. The parent cell would accumulate DNA and become polyploid with the potential occasionally to produce daughter bacteria containing DNA (see Figure 2). Alternately, if DNA replication preceded cytokinesis, then greater numbers of viable offspring would be produced at a faster rate. However, in this case once again DNA would accumulate in some bacteria due to unequal partitioning, leading to polyploidy. It is interesting to note that a population of both monoploid and polyploid bacteria seems to be a common outcome for all the predicted protocell division scenarios.
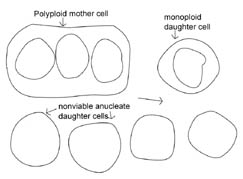
Figure 2. Hypothetical model of protocell division. In the protocell, DNA replication and cytokinesis would not be coordinated. Therefore DNA replication could occur faster or slower than cytokinesis. This figure shows a potential outcome of cytokinesis occurring at a faster rate than DNA replication. Note that many non-viable, anucleate daughter bacteria would be produced, as well as bacteria that are monoploid or polyploid. An animated version of this figure is available for viewing at: http://www.cedarville.edu/dept/sm/jwf/division.htm.
Since polyploidy is predicted to be a common outcome of protocellular life, and it since is generally detrimental to cellular life in contemporary cells, it is important to consider its effects on protocell evolution. Polyploidy would present at least two major hurdles for the evolving protocell. First, it would mean a diminished capacity for natural selection of favorable traits. For instance, Koch has calculated an upper limit for the number DNA copies the protocell could reasonably contain and still flourish. His upper limit for DNA is based on the fact that favorable mutations would be diluted in the “selfish” protocell carrying great numbers of chromosomes and duplicate genes (Koch, 1984).
Another problem that polyploid protocells would face is regulation of cell volume. The cell volume would eventually have to adjust to accommodate the increase in DNA and a corresponding increase in protein production. (In contemporary organisms that successfully harbor polyploid cells, those cells are larger than its typical cell.) How does the protocell adjust its size if there is no coordination between membrane events and biopolymer production? Perhaps the cell could inhibit uncontrolled DNA replication and protein accumulation by the production of an inhibitor of gene expression. But in that case, what would control the inhibitor so that it would not inhibit gene expression of all DNA, especially if the inhibitor is concentration-dependent and is transferred to a daughter cell with fewer DNA strands? We could postulate that an inhibitor would require a complex mechanism to ensure that gene expression from at least one DNA is not inhibited.
Despite these problems, is it possible that a viable bacterium with either a single DNA chromosome or several DNA chromosomes could be consistently produced throughout these reproduction cycles? Certain mathematical models show that this is possible, though production of nonviable cells would be common. It is also possible that this stochastic cell division process would create the condition we find in nature—namely, the continual production of bacteria that contain only one DNA molecule (monoploidy). However, the question remains how the unregulated division process of the protocell would lead to the highly organized and controlled division process that we observe in bacteria today.
To answer that question, we need to focus for a moment on those highly organized and regulated processes observed in the majority of bacteria today. Biochemical analysis reveals a bacterial cell division process that operates with remarkable precision. For example, E.coli bacteria replicate their DNA every 40 minutes (Lewin, 1997). In wild type E.coli, DNA replication and cytokinesis each occur at fixed time intervals (the latter takes 18 to 20 minutes), and the entire cycle is repeated with clockwork-like precision each time a bacterium divides. However, E.coli exposed to favorable conditions (when resources are plentiful) can divide as fast as 18 minutes, because the cell can overlap the fixed processes. That is, DNA replication can begin twice before the cell membrane divides, such that the new daughter bacteria receive DNA that is in the process of being replicated.
What is the signal for this increased rate of response? The trigger mechanism is unknown, but what is known is that bacterial cell division is coordinated precisely with the increase in bacterial cell mass. If the rate of cell growth is fast, the cell division mechanism responds by cycling at a faster rate. As a recent review article notes, “It is surprising that genetics, which has been a powerful tool in unraveling other regulatory circuits, has not yet been exploited to elucidate how E.coli regulates its mass” (Vinella, 1995). One theory suggests that as the cell increases its mass, it also increases the concentration of an “initiator” protein that triggers the cell cycle. Even though it is not yet clear how such an initiator may work, many of the events surrounding the initiation of both DNA replication and cytokinesis are being elucidated.
To better understand how both of these processes are coordinated and regulated during cell division, let’s take a look at what is known about the initiation of both of these events.
The goal of DNA replication in bacteria is the duplication of a circular DNA strand. Once DNA replication begins, the cell is committed to complete the process. The primary player in DNA replication is the DNA polymerase protein, which is a fairly large and complex protein that works in coordination with DNA to unwind proteins. Coordinating with it is a specialized ring-clamp protein that can literally glide up and down the DNA and help keep the DNA polymerase tethered to the DNA—an apparatus referred to as the replisome. How does the DNA polymerase “know” when and where to assemble the replisome?
Before the polymerase can find its start site, two problems must be solved: the DNA must unwind, and single-stranded DNA must be exposed. The unwinding and stabilization of single DNA strands involves an elaborate arrangement of proteins called an initiation complex, which assembles at a unique site on the DNA (see Figure 3). A protein designated DnaA is the most crucial protein involved in the initiation complex and it is found in many bacteria. DnaA recognizes specific nucleotide sequences and binds to them near a site on the DNA called the origin of replication. Individual DnaA proteins also have the special property of being able to bind to one another, and they exploit this ability by forming a cluster of up to 40 monomers, which causes the DNA to bend around the cluster. The bending stresses the structure of the double helix and several regions that are rich in A-T base pairs open up, exposing single strands of DNA. (A-T base pairs are weaker than G-C base pairs). However, the unwinding of the DNA causes tension in the DNA double helix, because the entire bacterial DNA chromosome is circular. This tension is relieved by two other proteins present in the initiator complex: DnaB and DnaC. This complex of DnaB and DnaC is so large that it appears as a blob when visualized with the aid of an electron microscope. It is often referred to as the engine of initiation.
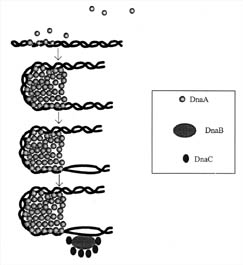
Figure 3. Initiation of DNA-replication in prokaryotes. DnaA protein binds to repeated sequences on the DNA near a site called the origin of replication. These repeated sequences are conserved and are called consensus sequences. The consensus sequences that the DnaA protein recognizes are of two classes, those containing nine nucleotides and those containing thirteen. The self-association of the DnaA monomers forms a cluster that causes the DNA to distort and bend. The bending causes unwinding of the DNA. The unwinding continues with the aid of three other proteins, Helicase (DnaB), DnaC, and gyrase. Single-stranded DNA is also stabilized by a protein called SSB (not shown). (Based on Figure 15.18 in Lewin, 1997.)
The initiator engine eventually displaces the DnaA and promotes the continued unwinding of DNA through the specific action of DnaB, which is a helicase. Helicases are ATP-dependent enzymes that break hydrogen bonds and can unwind DNA at a rate of 500-1000 base pairs per second. Helicases are classified as motor proteins, which are enzymes that convert chemical energy into physical movement. E.coli has 12 distinct helicases. The DnaB helicase works in conjunction with another protein called gyrase, which is part of another family of proteins essential for cellular life called topoisomerases. Gyrase has the remarkable ability to cut, unwind, and then rejoin DNA strands, relieving the tension created by the helicase-induced unwinding.
The DnaB helicase creates an interesting problem. The unwound single-stranded DNA is much less stable than the double-stranded form and it can potentially bind to itself. Another protein called single-stranded binding protein (SSB) binds to the strands and stabilizes them. The SSB proteins are required for replication, and a mutation in the gene for their production is lethal.
It certainly appears that several crucial factors must be in place for successful initiation of DNA replication, and moreover it appears that they work together in an irreducibly complex fashion.
A possible objection to the conclusion of irreducible complexity is that DNA replication can be performed in a cell-free system by adding back just a few components of the replication machinery. This procedure known as PCR (polymerase chain reaction) is a simple and powerful way to increase the concentration of isolated DNA in the laboratory. The procedure requires the use of DNA polymerase but does not require DnaA, helicase, or gyrase. One might conclude, therefore, that these components are not needed and that DNA replication can be achieved by a simpler, possibly non-irreducibly complex mechanism.
However this is not the case; the PCR reaction involves procedural steps that essentially replace the functions of the missing enzymes. For example, high temperature is used in the PCR reaction to unwind and “unzip” the DNA—essentially replacing the functions performed by the helicase and gyrase enzymes. Thus the same functions are always required for DNA replication, even if they are achieved in different ways.
Cytokinesis involves the coordination of many interacting components and must perform several major feats, including the synthesis of the membrane and cytoplasmic components required to create two bacteria from one. The synthesis of membrane and cell wall is quite an accomplishment considering the fact that many bacteria possess three outer layers: cytoplasmic membrane, cell wall, and outer membrane. During cell division, all three of these layers must be precisely extended in a short period of time, since production of daughter bacteria that are the same size and shape as the parent cell can occur rapidly.
Moreover, a majority of these new membrane and cell wall components are manufactured preferentially near the dividing point of the parent cell and are coordinated with constriction of the cell at the same location. The cytokinesis process also accurately partitions the DNA into each daughter cell, before the division of bacteria is completed. The DNA segregation mechanism is incredibly accurate, resulting in correct partitioning of the DNA greater than 99.9% of the time (Vinella, 1995).
The initiation of cytokinesis centers around a region on the membrane that will eventually become the dividing plane of the cell or the septum. Studies involving E.coli suggest that the septum is derived from a site on the membrane called the periseptal annulus. The periseptal annulus is a ring that encircles the cell and appears to be the result of an invagination or a melding of the inner membrane and the cell wall. The septum forms near this ring exactly at mid-cell. (It is unknown how the cell precisely measures the exact center of the cell.) Several proteins involved in septum formation form a complex at this site, working together to form a constriction ring, synthesize new membrane and cell wall, and break old membrane and wall attachments.
One of the earliest acting and most crucial proteins involved at this site is the FtsZ protein. The FtsZ protein self-polymerizes and is the primary component of a division ring that is hypothesized to constrict during cytokinesis. Upon completion of the septum, FtsZ depolymerizes. There is strong evidence to suggest that FtsZ is an essential cell division factor for free-living bacteria (Vincente and Errington, 1996). (Bacteria are considered to be free-living if they are capable of independent life free from a host organism or do not require complex nutritional factors that are typically supplied by the host organism.) The role of FtsZ in cell division is supported by the fact that it has been found in bacteria as diverse as mycobacteria and archaebacteria (Baumann and Jackson, 1996). Some studies report that it has some structural and functional similarities to the cytoskeletal proteins found in eukaryotes (Vincente and Errington, 1996). It also has the ability to self-polymerize into strands and cyclic structures in vitro (Erickson, 1996). The FtsZ protein concentration is regulated at the level of transcription and its concentration is estimated to be between 5000 and 20,000 molecules per cell. Some studies have suggested that the concentration of FtsZ fluctuates with the cell cycle and its concentration can change by as much as 50 percent.
Recent studies are uncovering a fascinating story about the biology of the FtsZ protein. The data show that FtsZ is regulated both temporally and spatially by transcription and inhibitor proteins. Most of the data for this system has been derived from studies of E.coli. The precise regulation of FtsZ is supported by several studies showing that a critical amount of FtsZ is needed for cell division, and that its overproduction or underproduction can cause cell division anomalies and affect viability (Lutkenhaus, 1993).
Let’s examine some of the proteins with which FtsZ interacts, several of which are located on the cell membrane at the mid-cell septum. Two proteins located at the E.coli septum, ZipA and FtsA, are required for proper FtsZ function and may be directly involved in regulating its actions (Ma, 1996). For instance, a regulatory role for FtsA is supported by the fact that a certain ratio of FtsZ/FtsA is required for cytokinesis to proceed. Any deviation from the critical FtsZ/FtsA ratio causes inhibition or alteration of cytokinesis.
There are several hypothetical models for how FtsZ could act to constrict the cell at the septum (Figure 4). One of the more intriguing aspects of FtsZ biology involves how it preferentially forms the constriction ring at the mid-cell septum. FtsZ has several other septal site choices for binding because multiple binding sites are present in a single bacterium. For instance, in a pre-division mother cell, new periseptal annuli are predicted to form very early on either side of the original annuli and are eventually placed at one quarter the length of the cell on each side of the mid-cell septum. These potential FtsZ binding sites will form the mid-cell septa in the newly formed daughter bacteria (see Figure 5). In addition, there are septum binding sites at the poles of the bacteria, since the poles were derived from septa from earlier cell divisions (see Figure 6).
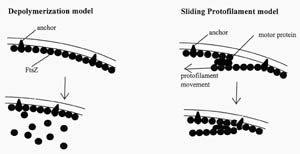
Figure 4. Two hypothetical models for how FtsZ could act at the septum. FtsZ is known to self-polymerize in vitro, forming rings and filaments. In the depolymerization model, FtsZ constricts the cell by polymerization-depolymerization cycles. In the sliding protofilament model, FtsZ protofilaments slide past one another. (Redrawn from Figure 8, Bramhill, 1997.)
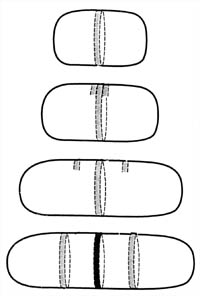
Figure 5. Derivation of the septum and periseptal annuli. The periseptal annulus, which is formed when the cytoplasmic membrane and the cell wall meld together, forms a ring around the cell precisely in the middle of the cell. It is hypothesized that the annulus serves as a site for development of the septum. New periseptal annuli have been detected forming very early in the life cycle of the bacterium and move away from the center annulus as the cell grows. They move to a position mid-way between the cell pole and the mid-cell annulus. They are then in position to serve as the mid-cell annulus of the newborn daughter bacteria.
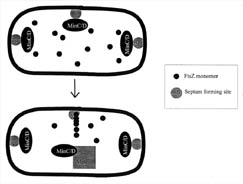
Figure 6. FtsZ is directed to the mid-cell septum by the combined actions of several proteins. Each bacterium has several septum locations where FtsZ can bind. Both poles of the bacteria contain septum complexes, since they were derived from mid-cell septa from their mother bacteria. FtsZ is directed toward the mid-cell septum region by the combined action of the MinC, D, and E proteins. The MinCD protein complex inhibits FtsZ binding at the poles and MinE overrides the inhibition at the mid-cell septum, allowing FtsZ to bind and polymerize. It is not known how MinE chooses the mid-cell septum.
How does the FtsZ protein discriminate between these sites? It is now believed that a set of proteins generated from a single genetic locus, the MinB locus, are involved in directing the FtsZ protein to the mid-cell septum. Three proteins from MinC, MinD, and MinE genes work together to inhibit cell division at the cell poles and promote it at the mid-cell septum. The data and current models show that the MinC and D proteins act together as an inhibitor and prevent the FtsZ protein from acting at any septal site. The MinE protein counteracts the effects of the MinC and D proteins precisely at the mid-cell, allowing the FtsZ protein to bind and polymerize there (see Figure 6). Apparently it is the ratio of MinE to MinCD that is important, since any deviation from an optimal MinCD/MinE ratio causes aberrant cell division. So, remarkably, FtsZ is controlled by the spatial concentration of Min proteins (Lewin, 1997).
Let’s consider the possibility that FtsZ and the Min proteins could have formed through natural selection. How does natural selection, using the random, uncontrolled division processes of the protocell, promote a cell division system that requires precise amounts of several essential factors in the right location at the right time? What is the selection pressure? Consider the FtsA, FtsZ, and Min proteins: each protein is a required component of the cytokinesis process; if one factor is missing, the cell does not divide properly. In fact, if the concentration of the factors is altered, cell division and cell viability are affected. Therefore, if evolution of all of the factors does not occur simultaneously, each factor alone could be a liability to the cell rather than an asset. If each of the factors alone is deleterious to life, then the evolution of each individual factor is less probable, since the cell lineage harboring the factor would tend to die out.
For instance, consider the scenario whereby evolution of the MinC, D, and E proteins occurred before evolution of FtsZ. Since it appears that one of the primary functions of the MinCDE system is to control FtsZ, what do the MinCDE proteins do after they evolve? We can postulate that the MinCDE proteins would be quite useless or even deleterious in the cell without the presence of FtsZ unless they were originally selected to perform another function.
Alternately, what if FtsZ evolved first? This seems more likely since FtsZ seems to be essential in all free-living bacteria. However, our current understanding is that FtsZ requires several binding proteins and the MinCDE system to successfully promote cell division at the mid-cell septum. Without the MinCDE proteins present in the cell, FtsZ will polymerize at the poles of the cell and cause the formation of anucleate mini-bacteria, diminishing the propagation of the cell lineage. This is supported by studies which have shown that if the FtsZ concentration is elevated in bacteria, it can overcome the MinCD mediated suppression of septation at the poles of the cell, and aberrant division can begin at several septal sites in the cell.
If FtsZ by itself has a negative or lethal effect on the propagation of cell lineages, could it have evolved in a dormant state before the evolution of its required co-factors? If so, what is the selection pressure that promotes the evolution of a dormant or inhibited FtsZ factor? The scientific evidence points to the fact that a MinCDE or equivalent system is required for FtsZ to function properly and supports the hypothesis that many factors would have to evolve rapidly and simultaneously for FtsZ-dependent cytokinesis to proceed. This seems to violate the basic tenants of Darwinian gradualism. Even if the MinCDE and FtsZ factors could have co-evolved we are still left with questions involving how the MinCDE system can select the mid-cell septum and how regulation of FtsZ polymerization occurs.
The fact that FtsZ requires several protein factors that work in a precise interdependent fashion to promote cytokinesis shows that the FtsZ-dependent cytokinesis mechanism present in E.coli is an irreducibly complex system. As such, it is highly questionable whether this complicated system could have arisen by Darwinian gradualism starting with a simple protocell.
Since it appears that it is unlikely that the MinCDE-FtsA-FtsZ-dependent cytokinesis apparatus found in E.coli could have existed in the early protocell, a biologist committed to philosophical naturalism could postulate that such a system may have evolved in the prebiotic soup, or else in the more stable late protocell.
First, let’s consider the derivation of the system by a gradual mechanism in the pre-biotic soup. This seems even more unlikely than its derivation in the early protocell. For instance, how could spatial control be achieved in the vast oceans of the prebiotic soup? Dilution would certainly be a problem. In addition, there is no membrane to divide, which is the primary reason for selecting such a system. Once again we can conclude that the spatial and temporal control of several factors involved in cytokinesis represents, at least in E.coli, a complex system that is difficult to account for by any gradualist theory.
By contrast, there is data that may support the evolution of the FtsZ-dependent cytokinesis system in the late protocell. For instance, even though the FtsZ protein is highly conserved, several bacteria lack some of the proteins that are part of the FtsZ-dependent cytokinesis system (see Table 1). Could these bacteria represent cells that have evolved only part of the FtsZ-dependent cytokinesis system or a simpler form of the system? Could these bacteria thus be close descendants of the late protocell?
|
Cytokenesis
Protein
|
|||||
| Bacterium |
FtsZ
|
FtsA
|
MinC
|
MinD
|
MinE/DivIV-A
|
| Escherichia coli |
x
|
x
|
x
|
x
|
x
|
| Synechocystis |
x
|
x
|
x
|
x
|
x
|
| Helicobacter pylori |
x
|
x
|
x
|
x
|
x
|
| Bacillus subtilis |
x
|
x
|
x
|
x
|
x
|
| Aquifex aeolicus |
x
|
x
|
x
|
x
|
x
|
| Thermotoga maritima |
x
|
x
|
x
|
x
|
|
| Archaeoglobus fulgidus |
x
|
|
|
x
|
|
| Borrelia burgdorferi |
x
|
x
|
|
x
|
|
| Pyroccocus horikoshii |
x
|
|
|
x
|
|
| Mycobacterium tuberculosis |
x
|
|
|
|
x
|
| Methanococcus jannaschii |
x
|
|
|
x
|
|
| Methanobacterium thermoautotrophicum |
x
|
x
|
|
x
|
|
| Treponema pallidum |
x
|
x
|
|
x
|
|
Table 1. Cytokinesis proteins present in the genomes of free-living bacteria as detected by amino acid sequence.
There is a one major problem with this suggestion. Proteins can be identified either by their function or by their amino acid sequence. Most of the missing cytokinesis proteins in these bacteria have been determined to be missing because their amino acid sequence is not found. But it is possible that a different protein is fulfilling the same role as the missing protein. For instance, the amino acid sequence of the MinE protein is not found in the bacterium Bacillus subtilis, but a protein designated DivIVA has been detected that fulfills the role of the MinE protein (Boche and Pichoff, 1998). Therefore, in B.subtilis the cytokinesis apparatus appears to be irreducibly complex even though it lacks MinE.
Using the Intelligent Design model, we could predict that since an irreducibly complex FtsZ-dependent cytokinesis mechanism exists in E.coli and B.subtilis, and since it appears to be essential to bacterial life, a similar system may exist in all bacteria. The components of the system could be different structurally but their functions would be the same or similar. At this point, however, since all the components have not been identified in all bacteria, all we can conclude is that the cytokinesis apparatus of E.coli fits the definition of an irreducibly complex system.
Since the complete genomes of several bacteria are known, it would be interesting to analyze these genomes by sequence analysis for the presence of components of the FtsZ-dependent cytokinesis apparatus. This would allow us to begin to determine which components exist and which have yet to be identified. This analysis would also help determine the minimal requirements needed for a cytokinesis apparatus and would represent a first step toward elucidating whether a simple or even non-irreducibly complex cytokinesis system exists.
FtsZ has been detected in all free-living bacteria analyzed for its presence. Based on data derived from the study of E.coli we will hypothesize that the proteins FtsZ, FtsA, MinC, MinD, and MinE/DivIVA represent a core cytokinesis apparatus. Table 1 shows which of these proteins have been detected by amino acid sequence analysis in the thirteen free-living bacteria whose complete genomes have been determined. Five of the thirteen bacteria species possess a full complement of these proteins. In the others, one or more of the proteins are missing. It is interesting to note that at least one Min protein is present with FtsZ in all the bacterial species. Could some of the Min proteins perform multiple roles, or is one Min protein sufficient? We could argue that perhaps the combination of FtsZ, one binding protein, and one septum-locator protein like MinE, represents a minimal irreducibly complex cytokinesis apparatus in bacteria. At least, so far the evidence points toward this conclusion. However some bacteria like mycoplasma, which often require a host cell and complex growth requirements, appear to be missing the amino acid sequence for a majority of these factors. Therefore, until all bacteria are analyzed, and cytokinesis proteins detected, we cannot make the claim that an irreducibly complex cytokinesis system is a universal phenomena in all free-living bacteria.
Even though a universal system has not yet been detected, genome analysis has revealed that the FtsZ protein itself is an important universal cytokinesis protein because it is found in all free-living and many non-free-living bacteria. This is intriguing because non-free-living bacteria, such as the mycoplasmas, often borrow proteins or energy from the host cell to survive and therefore they tend to have smaller genomes and fewer proteins, and yet mycoplasma possesses the FtsZ protein. However, Chlamydia trachomatis, a parasitic bacterium which requires another cell in order to grow and divide, is the first bacterium in which the FtsZ amino acid sequence has not been detected (Stephens et al., 1998). Even more interesting, the FtsA and MinD genes have been located in chlamydia. Could chlamydia harbor a cell division system that is FtsZ-independent, or a simpler cell division system? Could it even represent a transitional bacterium that has evolved only part of the cytokinesis apparatus?
Possibly. However, there is data to suggest that chlamydia use a complicated cell division mechanism and probably divide using a division ring like FtsZ. In fact, there are three lines of evidence supporting this. One line of evidence shows that chlamydia possess a cell division protein called cytoplasmic axial filament protein (cafa). This protein has been shown to be essential to division in some bacteria and may form filaments similar to FtsZ (Okada et al., 1994). Therefore, cafa could replace FtsZ. Second, non-free-living bacteria like chlamydia have been found to recruit cytoskeletal components from the cell they parasitize. One researcher suggests that the recruitment of cytoskeletal components could allow the bacteria to use the host cell proteins for stress fibers and cleavage rings (Rhee and Sanger, 1994; and Sanger, 1999). It is possible that chlamydia could operate in this manner. Third, cell division of chlamydia is affected (the cells increase in size) when the cells are exposed to cytoskeletal disrupting agents, suggesting that a cytoskeletal component is involved in division (Schramm and Wyrick, 1995). At this point, we will have to wait and see if other cytokinesis components will be discovered in chlamydia.
It also appears from sequence analysis that, in general, parasitic bacteria with small genomes, like mycoplasma, possess fewer of the core cytokinesis protein factors. However, it is also interesting to note that in the bacteria Aquifex aeolicus and Thermotoga maritima, two of the free-living bacteria considered by evolutionists to be the most ancient, an almost-full complement of the core cytokinesis factors are found (see Table 1). In fact, all the factors are found in Aquifex which has a genome one-third the size of E.coli.
FtsZ is speculated to play a wider role than just formation of the septum. For instance, it may play an important role in the timing of cytokinesis. If FtsZ has other functions and regulates other aspects of cell division, this would support the theory that it (or cytoskeletal proteins like it) may be essential to cell division.
To understand how FtsZ could play diverse roles in cell division we could speculate how it might regulate the timing of cell division with an increase in cell mass. We could hypothesize that daughter bacteria receive enough MinCD protein from the parent to maintain inhibition of division at all the septal sites of the cell. As the cell mass increases, so the does the concentration of FtsZ until it reaches its critical concentration. Since FtsZ is synthesized at a different rate from MinCD or FtsA, the correct ratio of FtsZ with these factors is eventually reached. This is supported by experimental evidence showing that cell division does not proceed unless FtsZ is in a certain proportion with MinCDE and FtsA. Once the required ratios are achieved, FtsZ polymerizes and, in coordination with MinE, causes formation of the division ring at the mid-cell septum.
This model is intriguing since it suggests a way in which mass could regulate division. However, it reveals nothing about how the division factors evolved; in fact, the model supports the contention that cytokinesis is a complex procedure involving many interdependent factors. In addition, the model does not yet account for how MinE actually promotes selection of the mid-cell septum. What it does imply, however, is that FtsZ could have diverse activities. This is supported by the fact that FtsZ works with several factors in addition to the ones we have mentioned. For instance, there are a number of factors in addition to FtsA and ZipA that operate at the mid-cell septum during cytokinesis and we will consider these next.
Several studies have shown that septum formation involves the coordinated interaction of several proteins for the production of new membrane and cell wall components (Bramhill, 1997). FtsZ is thought to interact with several of these proteins at the septum and is hypothesized to activate those involved in membrane and cell wall synthesis. Figure 7 shows the proteins known to be involved at the septum and their possible arrangement in the membrane. The number of components required and the functions that must be performed at the septum is impressive. The functions of the proteins and other factors required for cytokinesis are listed in Table 2. Many are critical to cell division and cellular life. The requirement of these factors and their regulation provides more evidence in support of the hypothesis that in E.coli and perhaps other bacteria, cytokinesis is an irreducibly complex system.
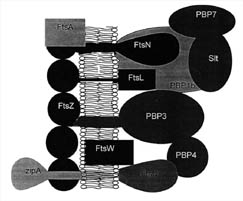
Figure 7. Proteins present at the mid-cell septum in some bacteria. FtsZ works in conjunction with several proteins at this site on the cytoplasmic membrane. FtsA and ZipA have been shown to be required for FtsZ function in E.coli. The Penicillin binding proteins (PBP) work together to synthesize cell wall components. The functions of several other proteins are described in Table 2. (Based on Figure 8, Bramhill, 1997.)
But FtsZ-dependent cell septum formation is not the only complex process involved in cytokinesis. Another remarkable process that occurs is the precise partitioning of DNA chromosomes to the new daughter bacteria. This is also known to be an active process dependent on several critical protein factors.
| Gene or site | Protein name and/or action |
| FtsZ | Essential cytoskeletal component |
| FtsA | Essential for FtsZ activity in E.coli, binds ATP |
| FtsK | Present at septum, may be involved in DNA segregation |
|
FtsL |
Transmembrane coordination |
| FtsN | Essential for division, periplasmic action |
| FtsQ | Periplasmic action at septum |
| FtsW | Located at septum PBP3 activator? |
| ZipA | Essential for FtsZ activity in E.coli, may be an anchor |
| PBP1b | Transpeptidase activity at septum |
| PBP3 | Penicillin-binding protein, Division specific transpeptidase (FtsI) |
| Slt | Lytic transglycolase |
| MinC | Division inhibitor |
| MinD | Enhances MinC inhibition activity |
| MinE | Site selection factor for MinC,D |
| SdiA | Transcription factor, activates FtsZ transcription |
| DnaK | Chaperone (HSP70) |
| GyrA GyrB |
DNA gyrases, unwind DNA |
| TopC TopE |
Topoisomerase IV, decatenates the chromosomes after DNA replication. |
| XerC XerD |
Recombinases that ensure fidelity of DNA recombination during DNA replication |
| MukB | Essential cytoskeletal protein involved in chro mosome partitioning |
| SpoOJ ParB |
Proteins involved in chromosome partitioning |
Table 2. Several of the proteins involved in cytokinesis
As we have noted, both monoploidy and polyploidy are likely karyotypic outcomes of early protocell evolution. Since monoploidy is the dominant form of genome structure found in contemporary bacteria, the protocell theory must eventually account for the dominance of the monoploid state in bacteria. Furthermore, in contemporary bacteria the monoploid state is not achieved by random partition processes but is postulated to involve an elaborate protein-dependent mechanism that results in accurate partitioning of DNA chromosomes greater than 99.9 percent of the time under optimal conditions. Recent evidence provides a remarkable picture of how this protein-driven partitioning mechanism is involved in specifically segregating bacterial DNA strands or chromosomes.
Three major problems must be solved by the partitioning mechanism if the DNA chromosomes are to be accurately distributed. (1) The circular DNA strands must be decatenated after DNA replication. Decatenation involves the unlinking of two circular DNA chromosomes that are catenated, which means they are linked together like two links of chain. (2) Once DNA is decatenated, there must be a mechanism to separate the strands and direct them to opposite poles of the mother cell. (3) The decatenation and separation of the DNA must occur before the septum wall forms and the daughter bacteria separate. Recent studies show that all three of these problems are overcome by an active protein-dependent partition process involving several proteins.
The partitioning process has been observed to begin soon after DNA replication initiation in both B.subtilis and E.coli (Levin and Grossman, 1998). The data supporting a partitioning process that begins very early in the cell cycle comes from the study of newborn daughter bacteria that have inherited a partially replicated DNA chromosome bearing two DNA replication origins. Using fluorescent tags attached to the origin regions, researchers have been able to observe the two replication origins being actively pulled apart, each toward a cell pole. As the cell cycle proceeds, the DNA finishes replication, is decatenated, and each new DNA strand moves towards its origin (see Figure 8). Each new daughter DNA then begins replication again in the mother cell before the completion of cytokinesis. The new replication origins are eventually polarized on each newly copied DNA strand such that one origin is directed toward the pole and one toward the forming septum of the mother cell (see Figure 8). When the cell divides, the two new daughter bacteria look similar to the parent cell, with the replication origins of the DNA oriented toward the poles of the newborn cell. In B.subtilis this partitioning phenomena is dependent on a protein produced by a gene called SpoOJ (Levin and Grossman, 1998). Mutations in SpoOJ can cause the formation of anucleate bacteria. SpoOJ protein is associated with the origin site on the DNA. It is postulated that SpoOJ could operate like a tether that actively segregates the DNA replication origins.

Figure 8. Active partitioning of the bacterial chromosome before completion of cytokinesis. DNA replication can occur very early in the cell cycle. The filled circles in the diagram represent proteins that have been observed binding on the DNA near the replication origin. (a) Researchers have noted that the origins are actively polarized toward opposite ends of the cell by a protein-dependent mechanism. (b) As the cell grows a septum begins to form and FtsZ begins to polymerize at mid-cell. (c) DNA replication is completed but begins again on each new DNA. (d) The replication origins are once again polarized by an active protein-dependent process such that each new daughter cell receives a single, partially replicated DNA chromosome with origins that are polarized towards the cell poles. (Drawing is adapted from Levin and Grossman, 1998.) A similar figure is available for viewing on the Internet in an animated form at http://www.cedarville.edu/dept/sm/jwf/division.htm.
This protein-dependent segregation mechanism has also been confirmed to exist in both Caulobacter crescentus and E.coli. A protein similar to SpoOJ has been identified in C. crescentus but has not been yet identified in E.coli. However, motor proteins that are involved in chromosome partitioning have been identified in E.coli and B.subtilis (Hiraga, 1993). In E.coli a protein produced by the mukB gene has been identified as a filamentous motor protein that binds to DNA (Figure 9). It is hypothesized that mukB may act like a cytoskeletal protein that actively moves the DNA, causing it to move toward the pole towards which the origin binding protein has been directed. The role of mukB in partitioning has been confirmed by observing bacteria with mutant mukB genes. The mutant bacteria frequently divide abnormally, producing anucleate bacteria and bacteria with two copies of DNA.
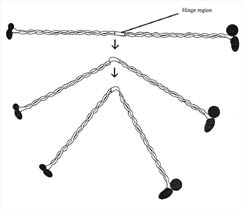
Figure 9. MukB protein. Drawing of the putative structure of the mukB motor protein. Each protein is made of several protein filaments and globular heads. The middle hinge region may allow mukB to bend. MukB is involved in the partitioning and packing of the bacterial chromosome during cell division. MukB has binding sites for both FtsZ and DNA. (Redrawn from Figure 1, Hiraga, 1993.)
Curiously, FtsZ mutants produce similar defects, suggesting that both FtZ and mukB protein may both be involved in partitioning. In fact, recent studies have shown that mukB can bind to FtsZ (Lockhart and Kendrick-Jones, 1998). These data suggest the exciting possibility that these proteins may interact to coordinate chromosome partitioning. Perhaps the relationship between mukB and FtsZ could account for the apparent coordination between septation and partitioning, by insuring that segregation occurs before septation. Thus, it appears that mukB and proteins like it could be another essential part of an irreducibly complex FtsZ-cytokinesis mechanism.
MukB is also known to act with several other proteins to package the DNA (Hiraga, 1993). This sounds like a simple feat but actually is a remarkable accomplishment, considering that the DNA is approximately 1000 times longer than the bacterium itself. In fact, each long circular DNA is packaged in a highly condensed state called the nucleoid. This means the cell must untangle and decantenate two very long rings of DNA, which involves untangling molecules that potentially have 200 or more folds (Dillon, 1981). The decatenation process involves topoisomerases that specifically perform the final separation process (see Figure 10). Additional proteins then participate in folding the DNA into its highly condensed nucleoid state so it can be packaged in the new daughter bacteria. Without the packaging proteins and the topoisomerases, the nucleoids do not separate, which can prevent cell division.
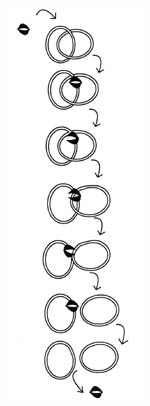
Figure 10. Decatentation of the replicated bacterial chromosomes by topoisomerases. After DNA is replicated, it is in a double-ringed, interlocked (catenated) form. There are several topoisomerases in the bacterial cell that maintain chromosome structure. In the case of decatenation, Topoisomerases IV binds to one of the DNA rings, cuts it, and allows passage of the other ring through.
Decatenation of the DNA rings may not be considered as complicated as some of the other processes, yet it is one of the most critical events of cytokinesis. If the DNA does not separate, production of two new daughter bacteria will not occur. Furthermore, imagine the tremendous potential for error involved in decatenation of DNA in a polyploid protocell. The problem of handling circular DNA was highlighted recently in a speculative review paper on microbial evolution, in which the authors challenge current dogma and claim that eukaryotes most likely evolved before prokaryotes, because eukaryotes have a much more unsophisticated system for replicating their genomes (primarily because the eukaryotic genome is linear and not circular) (Pennisi, 1998; and Jeffares, 1998).
Adding to the complexity of chromosome partitioning is the finding that bacterial DNA is maintained in a specific condensed arrangement in the nucleoid. The condensed state of the nucleoid is maintained by several protein factors, some of which are essential to bacterial life (see Table 3). In fact, studies are revealing that the nucleoid has a specific complex arrangement with the cell membrane. The interaction between the membrane and nucleoid structure is thought to create a channel-like substructure environment that harbors multi-enzyme complexes and even controls insertion of proteins into the membrane. A group of investigators are proposing that this complex substructure or “enzoskeleton” is a necessary organelle-like structure that is essential to bacterial life (Norris et al., 1996). Several other studies have shown that the interaction of factors that form the nucleoid enzoskeleton are essential to maintain its shape and may regulate transcription. Thus the nucleoid and its associated factors represent another potentially irreducibly complex system.
| Gene or site | Protein name and action |
| HNS | Nucleoid associated protein, a global transcription activator that controls over 30 proteins. Overexpression inhibits cell division. |
| HU | Promotes condensation of nucleoid and may control binding of other factors like gyrase. |
| SeqA | Proteins involved in sequestering the origin of replication HobH and preventing multiple re-initiations |
| Gyrase | Topoisomerase that induces negative supercoils |
| FIS | Protein that binds to nucleoid and may help it maintain its structure |
Table 3. Some of the proteins involved in maintenance of the nucleoid.
We have explored several basic and essential processes involved in bacterial cell division. It seems warranted to conclude that in some bacteria, both DNA replication and cytokinesis are irreducibly complex. The presence of these complex systems in bacteria—considered by many scientists to be “living fossils”—raises questions about the gradual derivation of such systems in the pre-biotic soup or the early protocell. Although the triggers and global regulators for these cell division processes have not been elucidated, intriguing new evidence shows the existence of factors coordinating activities such as DNA replication and cytokinesis. For instance, a “response regulator” protein called CtrA has been shown to regulate the cell cycle in C.crescentus by coordinating DNA replication with cell division. CtrA , a transcription factor, modulates the transcription of several cell-cycle promoters. Fascinatingly, CtrA is itself subject to temporal and spatial control by both phosphorylation and proteolysis (Shapiro, 1997). The presence of coordinating factors in bacteria like CtrA supports the idea that bacterial cell division is irreducibly complex.
In summary:
It is clear from these conclusions that cell division is not a trivial or simple biological mechanism. It will be exciting to see if an irreducibly complex cytokinesis apparatus is universal among bacteria. The existence of a universal irreducibly complex cytokinesis mechanism would challenge the validity of the protocell theory. Because cell division is a complex mechanism in many bacteria, it is reasonable to assume that even, in chlamydia and mycoplasma, a cytokinesis system exists and that it is irreducibly complex. The existence of several different kinds of irreducibly complex cytokinesis apparatuses (e.g., one that is FtsZ-independent) would be even more problematic for protocell theorists to explain. Future studies that focus on identifying cytokinesis factors by functional analysis will be very helpful in determining the nature of the complexity of this process. For instance, analysis of chlamydia for the presence of an endogenous FtsZ-like protein, or one recruited from the host cell, will help determine if a division ring protein is essential for division.
Acknowledgments: This project was supported by awards from the Faculty Summer Scholarship Program and the Computer Services Faculty Incentive Fund at Cedarville College.
1. The focus of this paper is on the ability of biopolymers (proteins and nucleic acids) and bio-synthetic pathways to work together in a coordinated fashion to create the intricate clockwork-like conditions that promote cell division. I do not address the question of whether these biopolymers themselves could have originated by a gradualistic, evolutionary process in a prebiotic soup. This problem has been addressed in several reviews (Swee-Eng, 1996, and Mills, 1996).
Baumann P., and Jackson S. “An archaebacterial homologue of the essential eubacterial cell division protein FtsZ.” Proceeding of the National Academy of Science 93 (1996):6726-6730.
Behe, Michael. Darwin’s Black Box. New York: The Free Press, 1996.
Bouche, J.P., and Pichoff, S. “On the birth and fate of bacterial division sites.” Molecular Microbiology 29 (1998):19-26.
Bramhill, D. “Bacterial cell division.” Annual Review of Cell and Developmental Biology 13 (1997):395-424.
Cooper, Geoffrey. The Cell: A molecular Approach. Washington DC:ASM Press, 1997.
Dillon, S. Ultrastructure, Macromolecules and Evolution. New York: Plenum Press, 1981.
Erickson H., et al. “Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers.” Proceeding of the National Academy of Science 93 (1996):519-523.
Hiraga, S. “Chromosome partition in Escherichia coli.” Current Biology 5 (1993):789-801.
Javor, George. “Life an evidence for creation.” Proceedings of the Fourth International Conference on Creationism. Pittsburgh: Creation Science Fellowship, 1998.
Jeffares, D. “Relics from the RNA world.” Journal of Molecular Evolution 46 (1998):18-36.
Koch, Arthur. “Evolution vs. the number of gene copies per primitive cell.” Journal of Molecular Evolution 20 (1984):71-76.
Levin, P., and Grossman, A. “Cell cycle: the bacterial approach to coordination.” Current Biology 8 (1998):R28-R31.
Lewin, Benjamin. Genes VI. New York: Oxford University Press. 1997.
Lockhart A, and Kendrick-Jones J. “Interaction of the N-terminal domain of mukB with the bacterial tubulin homologue FtsZ.” FEBS Letters 430 (1998):278-282.
Loomis, William. Four Billion Years. Sunderland Mass: Sinauer Associates, 1988.
Lutkenhaus, J. “Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring.” Journal of Bacteriology, 175 (1993):1118-1125.
Lutkenhaus, J. “Escherichia coli cell division.” Current opinion in genetics and development 3 (1993):783-788.
Ma, X. et al. “Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli bacteria by using green fluorescent protein.” Proceeding of the National Academy of Science 93 (1996):12998-13003.
Mills, G. and Kenyon D. “The RNA world: A critique.” Origins and Design. 17 (1996):9-14.
Morowitz, Harold. Beginning of Cellular Life. New Haven and London: Yale University Press, 1992.
Norris, V., Turnock, G., and Sigee, D. “The Eschericia coli enzoskeleton.” Molecular Microbiology 19 (1996):197-204.
Okada Y., et al. “Possible function of the cytoplasmic axial filaments in chromosome segregation and cellular division of Eschericia coli.” Science Progress 77 (1994):253-264.
Pennisi, E. “Direct descendants from an RNA world.” Science 280 (1998):673.
Rhee, D. et al. “The premyofibril: evidence for its role in myofibrillogenesis.” Cell Motility and Cytoskeleton 28 (1994):1-24.
Sanger, Joseph. Personal web page. Retrieved Feb 15, 1999 from the world wide web (http://www.med.upenn.edu/~ifem/js.htm).
Schramm, N., and Wyrick, P.B. “Cytoskeletal requirements in Chlamydia trachomatis infection of host cells.” Infection and Immunity 63 (1995):324-332.
Shapiro, L., and Losick, R. “Protein localization and cell fate in bacteria.” Science 276 (1997):712-718.
Stephens, R.S. “Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis.” Science 282 (1998):754-759.
Swee-Eng, A. “The Origin of Life: A critique of current scientific models.” Creation Ex Nihilo Technical Journal 10 (1996):300-314.
Vincente, M., and Errington, J. “Structure, function and controls in microbial division.” Molecular Microbiology 20 (1996):1-7.
Vinella, D. and D’Ari, R. “Overview of controls in the Escherichia coli cell cycle.” BioEssays 17 (1995):527-536.
Copyright 2000 Access Research Network. All rights reserved.
International copyright secured.
File Date: 6.27.01